CellFree Sciences
"It is our goal to make proteins work!" CellFree Sciences is using its own wheat germ cell-free protein expression system to serve protein needs in life sciences and for industrial use. Performing protein synthesis outside of the cell in an in vitro system allows us to separate protein production from other processes required to maintain a cell. Thus, working cell-free opens new opportunities to better adopt protein synthesis to protein needs offering dedicated reagents to make for example membrane proteins in a lipid environment, labeled proteins for use in MS or NMR, or well-folded antigens for antibody production. Our scalable and flexible expression system can serve protein needs from the small amounts used for production of our protein bead arrays presenting up to 23,000 human full-length proteins to individual proteins made on large-scale for use in structural biology, medical research, on-demand product production, and diagnostic tests. Our eukaryotic expression system has a strong record on supporting a wide range of projects working on all kinds of proteins from the kingdoms of life.
View all CellFree Sciences Products
Wheat Germ Cell-Free Protein Expression System
The Power of Wheat Driving Science
CellFree Sciences is using its own wheat germ cell-free protein expression system to serve protein needs in life sciences and for industrial use. Performing protein synthesis outside of the cell in an in vitro system allows us to separate protein production from other processes required to maintain a cell.
Features
- High-performance wheat germ extracts developed at CFS for superior protein yields
- Dedicated extracts for expression of GST-tagged and His-tagged proteins (G and H versions)
- Dedicated amino acid-free reagents for protein labelling (WEPRO®8240 Series)
- Examples show the expression of DHFR using 6 ml bilayer reactions on CFS’ fully automated Protemist DTII protein synthesizer.
- Precleared wheat germ extracts of the G or H versions allow for higher protein purity after one-step protein purification.
Supported Reaction Formats
- ONE Kit: Coupled transcription-translation system for quick protein expression experiments
- Protein Research Kits: Premixed transcription and translation reagents to perform bilayer reactions
- Core Kits: Individual reagents to perform bilayer reactions on different scales
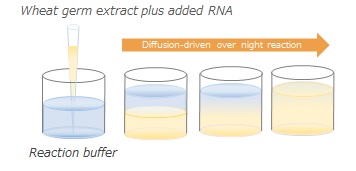
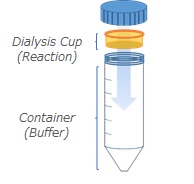
- Dialysis Kits: Individual reagents to perform dialysis-driven expression reactions
Special Applications
- FLEXIQuant standards: Modified reaction conditions allow for up to 99% labelling efficiency for incorporating [13C, 15N]-L-Lys and [13C, 15N]-L-Arg from Cambridge Isotope Laboratories to prepare standards for quantitative protein MS, studies on PTMs, and proteomics
- NMR samples: Expression kits with isotope-labelled amino acids from Cambridge Isotope Laboratories
- ProteoLiposome BD Kit: Expression of membrane proteins in the presence of added liposomes